rpoC Database

rpoCdb is an interactive reference database designed for high-resolution microbiome profiling that provides:
- Curated, high-quality full-length rpoC gene sequences
- Pre-formatted databases for commonly used microbiome software analysis pipelines
- Optimized wet-lab protocols
- Web-based taxonomy browser
Mutational Distance from Modal Haplotype Calculator
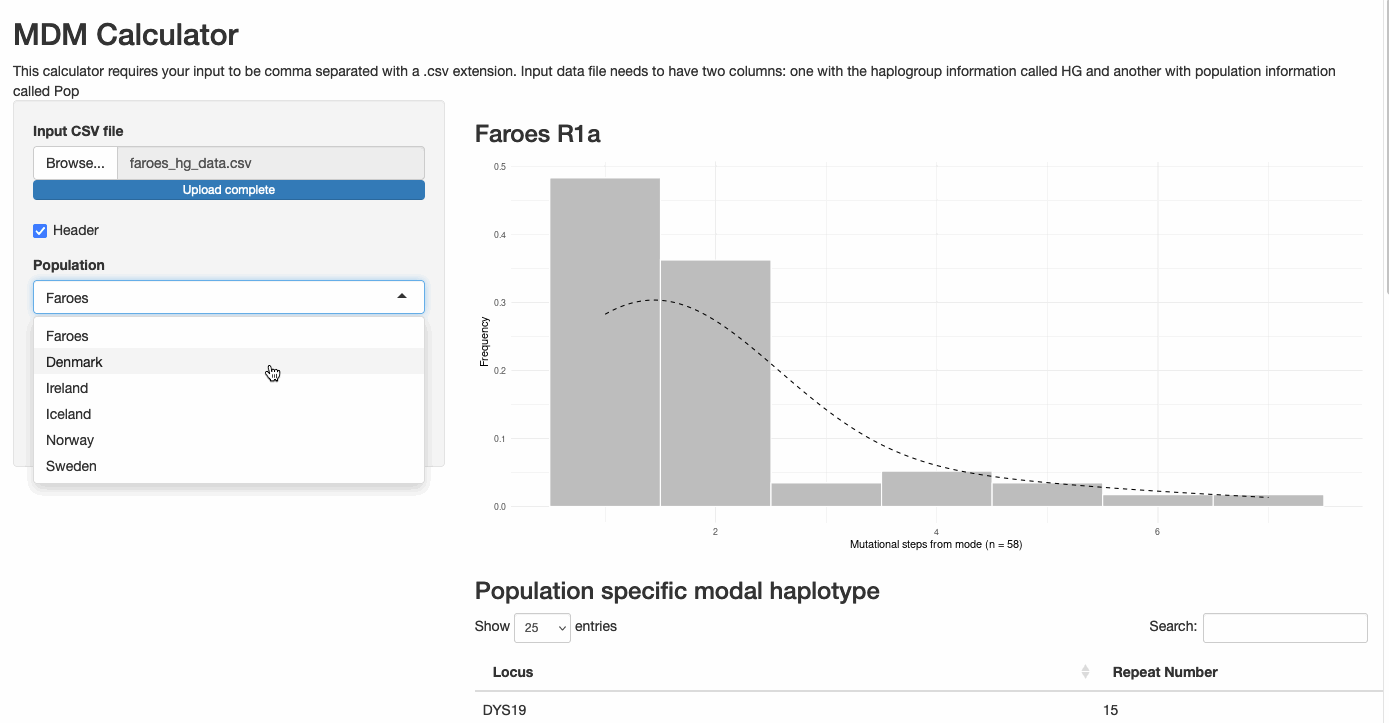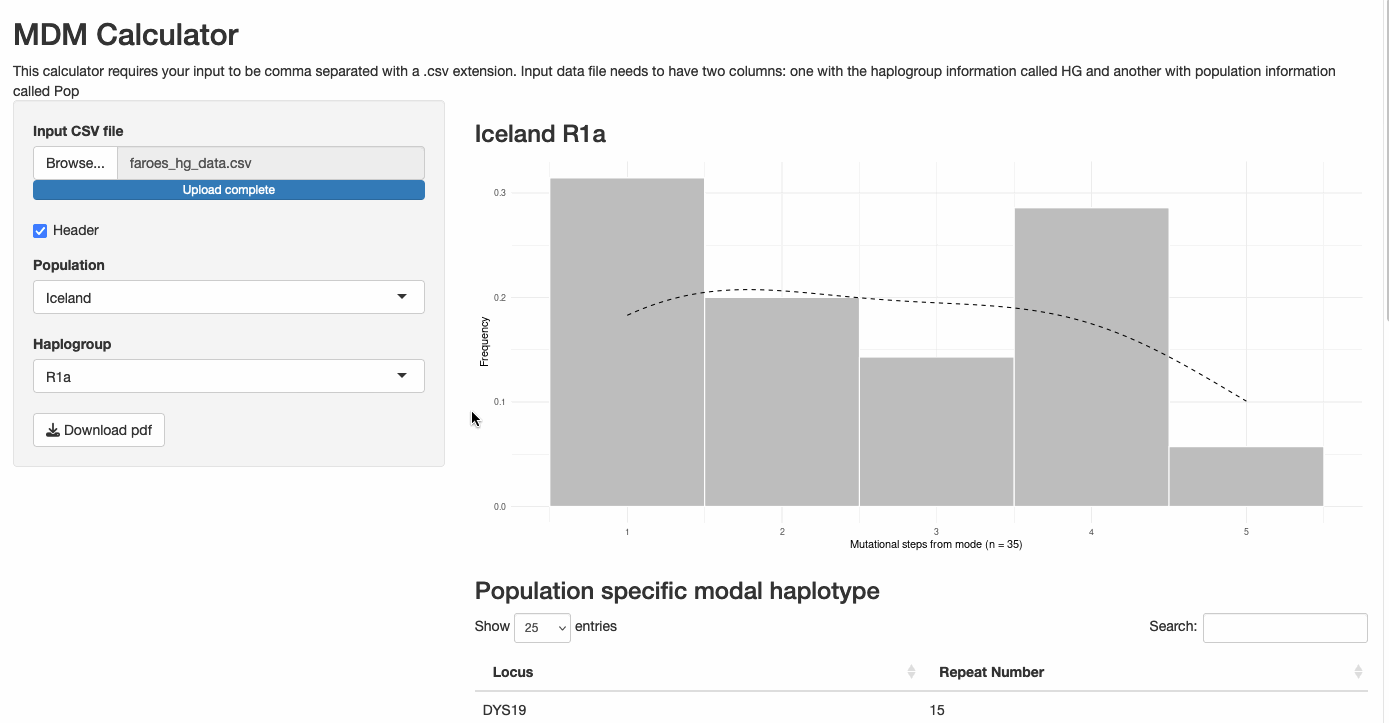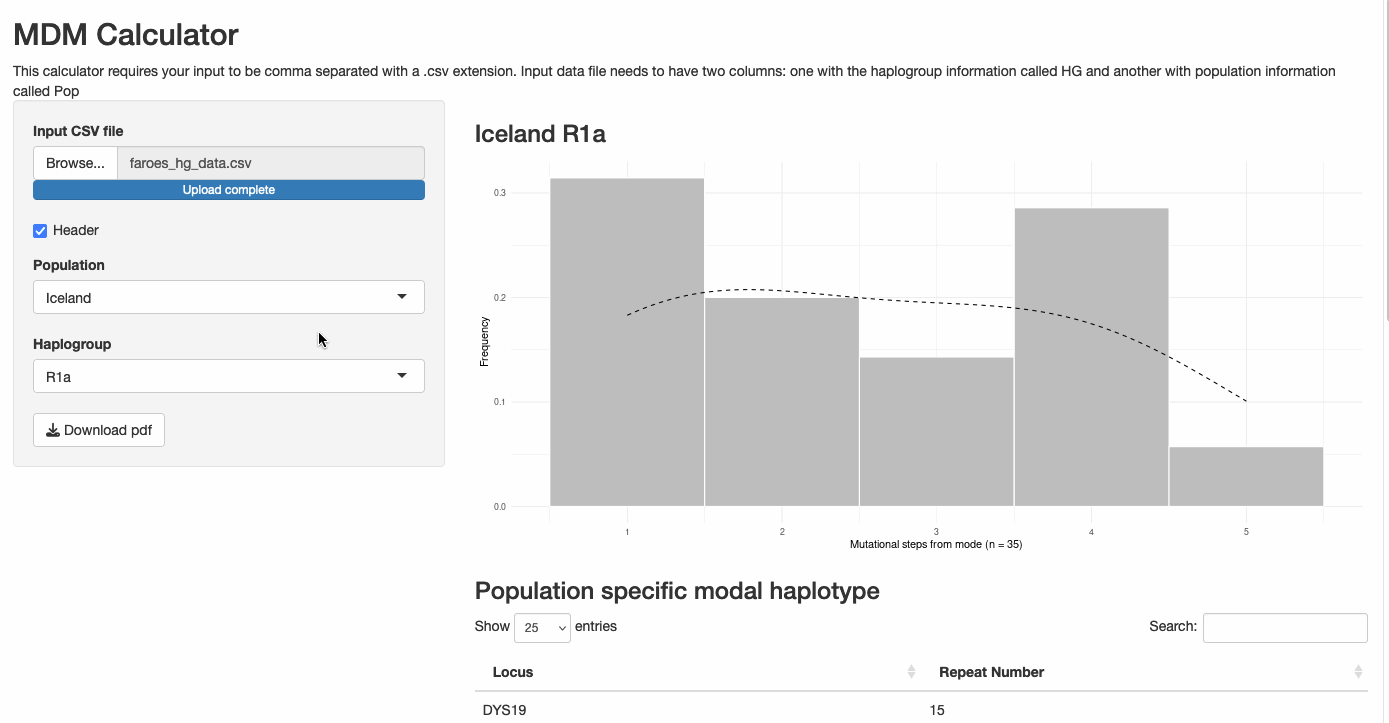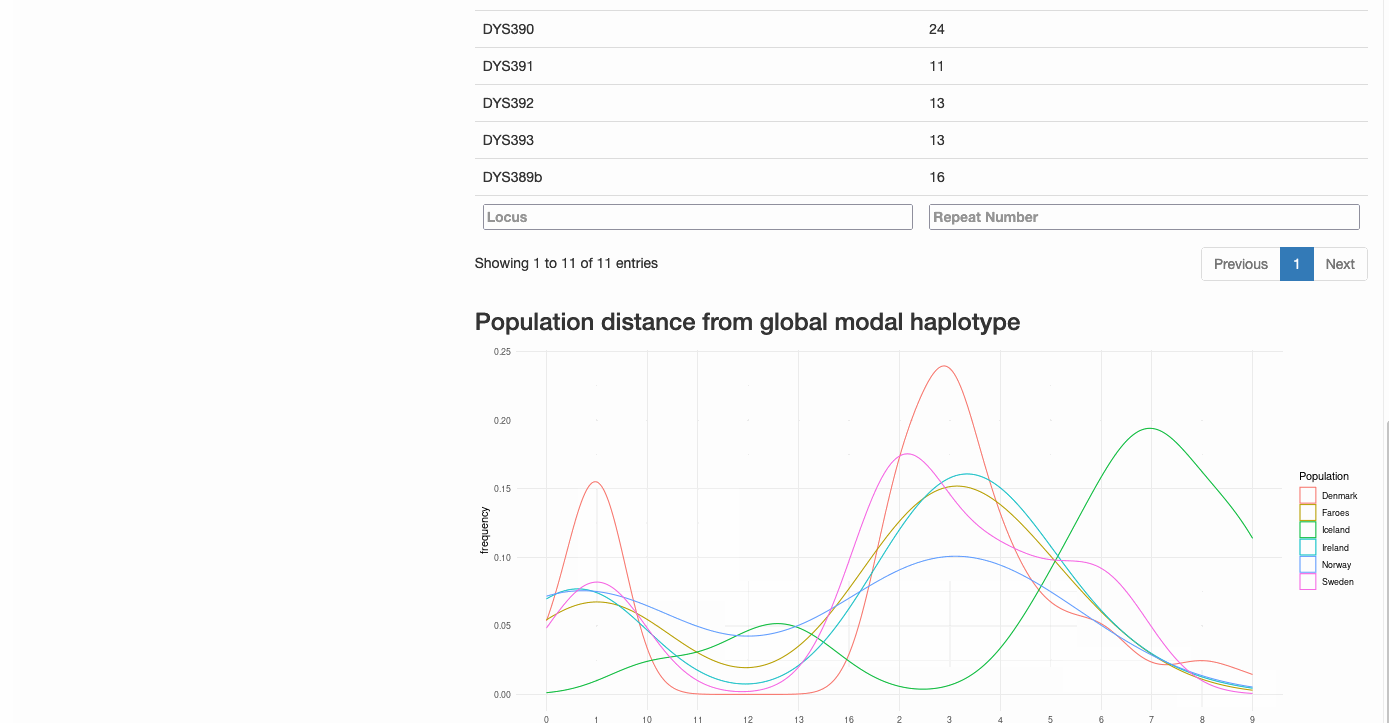
This interactive Shiny application uses STR repeat data from Y-chromosomal haplotypes to:
- Calculate the modal haplotype from input data
- Compute mutational distances for each individual from the modal haplotype
- Generate publication-quality visualizations of the distribution
- Provide statistical insights into population genetic patterns
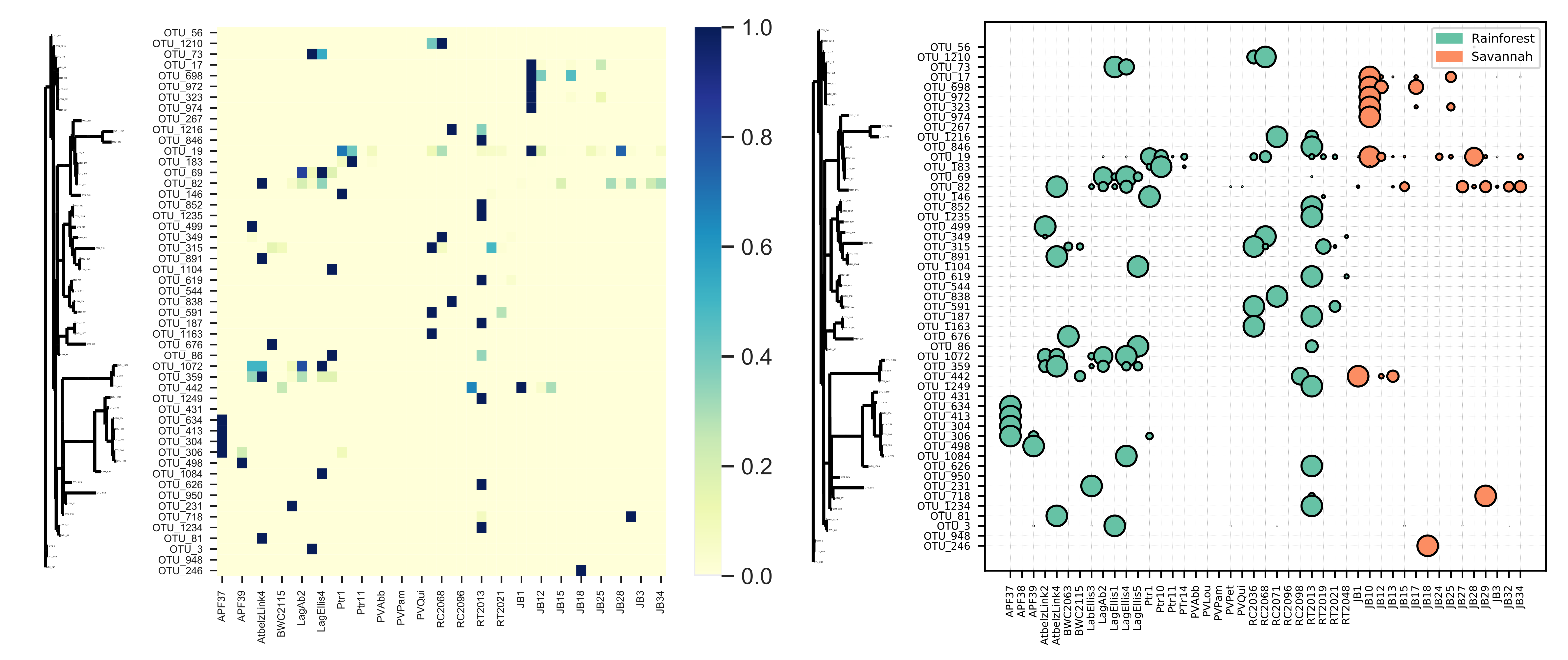
Bubble Tree
Bubble Tree is a visualization tool that integrates:
- Taxa frequency tables (rows as taxa, columns as samples)
- Phylogenetic tree structures
- Sample metadata
to create either bubble tree visualizations or heatmaps displaying abundances plotted on phylogenetic trees.
 View on GitHub
Conda Package
View on GitHub
Conda Package
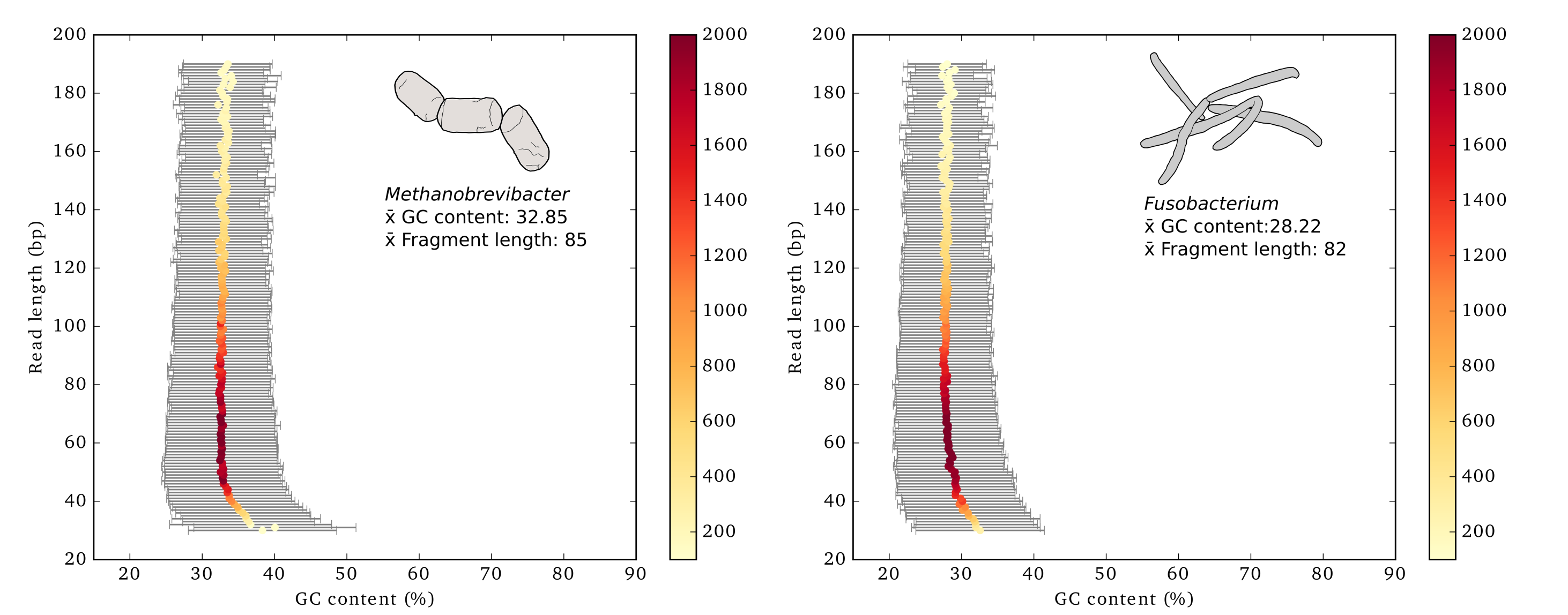
GC Content by Read Length Analysis
This tool provides comprehensive analysis of sequencing data by:
- Calculating summary statistics for read length and GC content from FASTQ/FASTA files
- Generating detailed statistics tables
- Creating publication-ready visualizations of GC content by read length
 View on GitHub
View Published Paper
View on GitHub
View Published Paper